Superoxide O
2•-, a reactive oxygen species, is a free radical produced in living cells that use oxygen. This radical can be toxic to living organisms by reacting with DNA or disrupting protein production. Superoxide dismutases (SOD) are metalloenzymes that catalyze the dismutation of superoxide. This reaction takes place at their active site based on metal ions (Fe, Mn, Cu, Zn). Recent studies have focused on a bacterial SOD, NiSOD, whose active site has a nickel ion. The understanding of the catalytic mechanism of NiSOD and the optimization of bio-inspired complexes of this enzyme could lead to the design of new therapies against oxidative stress related diseases, such as chronic inflammatory bowel diseases
In some bacteria, the dismutation of superoxide O
2•- is catalyzed by the enzyme NiSOD which active site is a square planar complex of the metal ion NiII. In order to elucidate the catalytic mechanism of this enzyme (
Figure 1), researchers at our laboratory and the Department of Molecular Chemistry DCM at UGA are developing complexes inspired by the active center of the NiSOD, by varying the coordination of the nitrogen and sulfur atoms around the Ni
II ion. A tripeptide derived from the ATCUN (Amino Terminal Cu
II and Ni
II) binding motif reproduces the N
3S
1 coordination of the metal ion in a square planar geometry (
Figure 2). Under physiological conditions (water, pH 7), the results obtained with this complex show a SOD-like catalytic activity. Despite their short lifetime and low concentration, the researchers could identify, in organic solvent and at low temperature, two unstable reaction intermediates: the superoxo Ni
II, and hydroperoxo Ni
II complexes (
Figure 2). The latter shows a direct interaction of the superoxide with the metal ion, indicating an inner sphere mechanism during catalysis. These two complexes characterized experimentally by Electronic Paramagnetic Resonance and by spectroelectrochemistry have signatures in agreement with the calculations obtained by DTF (Density Functional Theory).
These bio-inspired complexes constitute versatile structures that have the advantage of being stable in water and in organic solvents and provide insight into the mechanisms of superoxide dismutation by the NiSOD enzyme. In the future, researchers will optimize these bio-inspired complexes in the hope of developing new molecules to fight the harmful effects of oxidative stress.
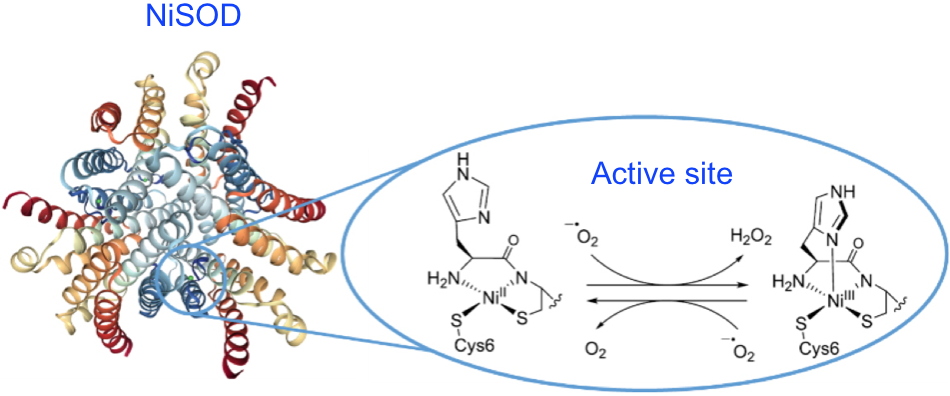
Figure 1: At the core of the NiSOD superoxide dismutase active site (
pdb 1T6U), the square planar Ni
II center is coordinated to two cysteinate thiolate functions, the terminal amine of the peptide chain and an amidate function of a peptide bond. The Ni
III state formed during dismutation has a square-based pyramid-like geometry due to the additional bonding of the imidazole of the histidine in the axial position.
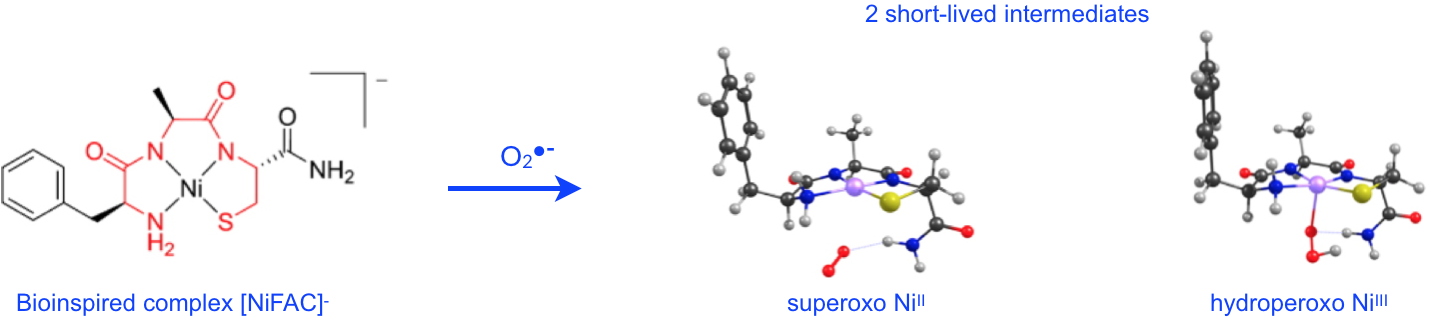
Figure 2: Upon reaction with superoxide, the bio-inspired NiSOD complex, [NiFAC]
- reveals two short-lived reaction intermediates:
1) the superoxo Ni
II complex involves binding of O
2•-via a terminal NH
2 group;
2) the hydroperoxo Ni
III complex shows the direct interaction of O
2•- with the nickel ion.
The structures are optimized by DFT: C (black), H (grey), O (red), N (blue), S (yellow), Ni (pink).
Project supported by the EUR-CBH (Graduate School of Chemistry, Biology and Health).